代表论文:
# 同等贡献,* 通讯作者
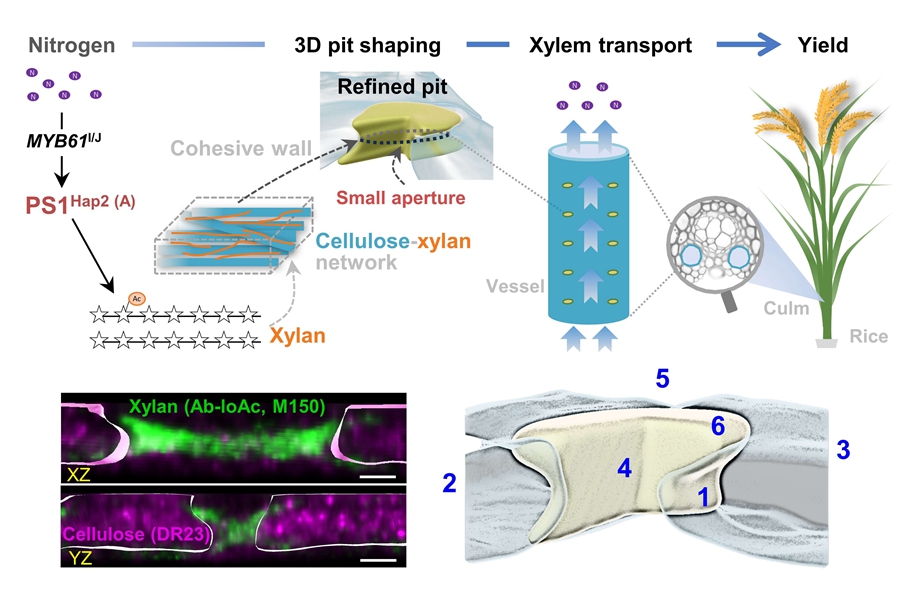
1. Zhang L#, Gao Y#, Xu Z, Tian Y, Hu J, Wen Z, Li J, Gao C, Zhou Y*, Zhang B*. (2025) Shaping pit structure in vessel walls sustains xylem hydraulics and grain yield. Cell 188: 7238–7251.e15.
2. Zhang L, Gao C, Gao Y, Yang H, Jia M, Wang X, Zhang B*, Zhou Y*. (2025) New insights into plant cell wall functions. J Genet Genomics 52: 1308–1324.
3. Wen Z#, Xu Z#, Zhang L#, Xue Y#, Wang H, Jian L, Ma J, Liu Z, Yang H, Huang S, Kang X, Zhou Y, Zhang B. (2025) XYLAN O-ACETYLTRANSFERASE 6 promotes xylan synthesis by forming a complex with IRX10 and governs wall formation in rice. Plant Cell 37: koae322.
4. Zhang L, Zhou Y, Zhang B*. (2024) Xylan-directed cell wall assembly in grasses. Plant Physiol 194: 2197–2207. (Invited review)
5. Xu R#, Liu Z#, Zhou Y, Zhang B*. (2024) Xylan clustering on the pollen surface is required for exine patterning. Plant Physiol 194: 153-167. (Invited research article)
6. Zhou Y#, Gao Y#, Zhang B#, Yang H, Tian Y, Huang Y, Yin C, Tao J, Wei W, Zhang W, Chen S, Zhou Y*, Zhang J*. (2024). CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice. Plant Cell 36: 3751–3769.
7. Cao S, Wang Y, Gao Y, Xu R, Ma J, Xu Z, Shang-Guan K, Zhang B*, Zhou Y* (2023) The RLCK-VND6 module coordinates secondary cell wall formation and adaptive growth in rice. Mol Plant 16: 999-1015.
(Recommended by H1 Connect)
8. Xu Z#, Gao Y#, Gao C#, Mei J, Wang S, Ma J, Yang H, Cao S, Wang Y, Zhang F, Liu X, Liu Q, Zhou Y*, Zhang B*. (2022) Glycosylphosphatidylinositol anchor lipid remodeling directs proteins to the plasma membrane and governs cell wall mechanics. Plant Cell 34: 4778–4794.
(Highlighted by The Plant Cell 2022, 34: 4671–4672)
9. Wang H#, Yang H#, Wen Z, Gao C, Gao Y, Tian Y, Xu Z, Liu X, Persson S, Zhang B*, Zhou Y*. (2022) Xylan-based nanocompartments orchestrate plant vessel wall patterning. Nat Plants 8: 295–306.
(Highlighted by Research Briefing in Nature Plants 2022, 8:330–331)
10. Zhou Y, Zhang B. (2022) Unprecedented polysaccharide nanostructures sustain vessel wall patterning and robustness. Nat Plants 8: 330–331. (Research Briefing)
11. Peng J#, Zhang B#, Chen H#, Wang M#, Wang Y, Li H, Cao S, Yi H, Wang H, Zhou Y*, Gong J*. (2021) Galactosylation of rhamnogalacturonan-II for cell wall pectin biosynthesis is critical for root apoplastic iron reallocation in Arabidopsis. Mol Plant 14: 1640–1651.
12. Zhang B, Gao Y, Zhang L, Zhou Y. (2021) The plant cell wall: biosynthesis, construction, and functions. J Integr Plant Biol 63: 251–272. (Invited review)
14. Gao Y#, Xu Z#, Zhang L#, Li S, Wang S, Yang H, Liu X, Zeng D, Liu Q, Qian Q, Zhang B*, Zhou Y*. (2020) MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat Commun 11: 5219.
15. Xue J
#,
Zhang B#, Zhan H
#, Lv Y, Jia X, Wang T, Yang N, Lou Y, Zhang Z, Hu W, Gui J, Cao J, Xu P, Zhou Y
, Hu J, Li L*, Yang Z*. (2020) Phenylpropanoid derivatives are essential components of sporopollenin in vascular plants. Mol Plant 13: 1644–1653.
16. Zhang L, Zhang B*, Zhou Y*. (2019) Cell wall compositional analysis of rice culms. Bio-protocol 9: e3398.
17. Wang S#, Yang H#, Mei J, Liu X, Wen Z, Zhang L, Xu Z, Zhang B*, Zhou Y*. (2019) A rice homeobox protein KNAT7 integrates the pathways regulating cell expansion and wall stiffness. Plant Physiol 181: 669–682.
(Highlighted by Plant Physiology 2019, 181: 385–386)
18. Zhang L, Gao C, Mentink-Vigier F, Tang L, Zhang D, Wang S, Cao S, Xu Z, Liu X, Wang T, Zhou Y*, Zhang B*. (2019) Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. Plant Cell 31: 1113–1126.
(Highlighted by The Plant Cell 2019, 31: 936)
19. Zhang Z
#,
ZhangB#, Chen Z, Zhang D,
Zhang
H, Wang H, Zhang Y, Cai D, Liu J, Xiao S, Huo Y, Liu Jie, Zhang L, Wang M, Liu X,
Xue Y, Zhao L*, Zhou Y*, Chen H*. (2018) A
PECTIN METHYLESTERASE at the Maize
Ga1 locus confers male function in unilateral cross-incompatibility.
Nat Commun 9: 3678.
20. Zhang D, Xu Z, Cao S, Chen K, Li S, Liu X, Gao C, Zhang B*, Zhou Y*. (2018) An uncanonical CCCH-tandem zinc finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol Plant 11: 163–174.
21. Zhang B#, Zhang L#, Li F#, Zhang M, Liu X, Wang H, Xu Z, Chu C*, and Zhou Y*. (2017) Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat Plants 3: 17017.
(Highlighted by Nature Plants 2017, 3: 17017)
22. Gao Y#, He C#, Zhang D, Liu X, Xu Z, Tian Y, Liu XH, Zang S, Pauly M, Zhou Y*, Zhang B*. (2017) Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol 173: 470–481.
23. Shi Y, Liu X, Li R, Gao Y, Xu Z, Zhang B*, Zhou Y. Retention of OsNMD3 in the cytoplasm disturbs protein synthesis efficiency and affects plant development in rice. (2014) J Exp Bot 65: 3055–3069.
24. Wu B#, Zhang B#, Dai Y#, Zhang L, Shang-Guan K, Peng Y, Zhou Y*, Zhu Z*. (2012) Brittle Culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol 159: 1440–1452.
(Recommended by F1000 Prime)
25. Ning J#, Zhang B#, Wang N, Zhou Y*, and Xiong L*. (2011) Increased leaf angle1, a Raf-Like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of rice. Plant Cell 23: 4334–4347.
26. Zhang B#, Liu X#, Qian Q#, Liu L, Dong G, Xiong G, Zeng D, and Zhou Y. (2011) A Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc Natl Acad Sci USA 108: 5110–5115.
(Recommended by F1000 Prime)
27. Zhang B and Zhou Y. (2011) Study on rice brittleness mutants: A way to open the ‘black box’ of monocot cell wall biosynthesis. J Integr Plant Biol 53: 136–142.
28. Zhang M#, Zhang B#, Qian Q#, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y. (2010) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63: 312–328.
29. Zhang B#, Deng L#, Qian Q#, Xiong G, Zeng D, Li R, Guo L, Li J, Zhou Y. (2009) A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol Biol 71: 509–524.
合作研究论文:
Ge F, Wei H, Zhang W, Mei C, Gao Y, Yang H, Wang X, Cao J, Liu Z, Shi J, Xia R, Zhao K, Tang S, Teng K, Zhong J, Yang F, Zhang B, Zhou Y, Yu F, Wu Y, and Xie Q. (2025) Vascular-tissue-specific expression of an endo-1,4-β-xylanase enhances forage efficacy of sweet sorghum silage. Mol Plant 18: 1241–1244.
Li W, Lin Y, Chen Y, Zhou C, Li S, Ridder N, Oliveira D, Zhang L, Zhang B, Wang J, Xu C, Fu X, Luo K, Wu A, Demura T, Lu M, Zhou Y, Li L, Umezawa T, Boerjan W, Chiang V. (2024) Woody plant cell walls: Fundamentals and utilization. Mol Plant 17: 114–142.
Renou J, Li D, Lu J, Zhang B, Gineau E, Ye Y, Shi J, Voxeur A, Akary E, Marmagne A, Gonneau M, Uyttewaal M, Höfte H, Zhao Y, Vernhettes S. (2024) A cellulose synthesis inhibitor affects cellulose synthase complex secretion and cortical microtubule dynamics. Plant Physiol 196: 124–136.
Chen C, Zhang Y, Cai J, Qiu Y, Li L, Gao C, Gao Y, Ke M, Wu S, Wei C, Chen J, Xu T, Friml J, Wang J, Li R, Chao D, Zhang B, Chen X, Gao Z. (2023) Multi-copper oxidases SKU5 and SKS1 coordinate cell wall formation using apoplastic redox-based reactions in roots. Plant Physiol 192: 2243-2260.
Li F, Yang J, Sun Z, Wang L, Qi L, A S, Liu Y, Zhang H, Dang L, Wang S, Luo C, Nian W, O’Conner S, Ju L, Quan W, Li X, Wang C, Wang D, You H, Cheng Z, Yan J, Tang F, Yang D, Xia C, Gao G, Wang Y, Zhang B, Zhou Y, Guo X, Xiang S, Liu H, Peng T, Su X, Chen Y, Ouyang Q, Wang D, Zhang D, Xu Z, Hou H, Bai S, Li L. (2023) Plant-on-chip: Core morphogenesis processes in the tiny plant Wolffia australiana. PNAS Nexus 2: 1-18.
Wang Y, Zou Y, Chen X, Li H, Yin Z, Zhang B, Xu Y, Zhang Y, Zhang R, Huang X, Yang W, Xu C, Jiang T, Tang Q, Zhou Z, Ji Y, Liu Y, Hu L, Zhou J, Zhou Y, Zhao J, Liu N, Huang G, Chang H, Fang W, Chen C, Zhou D. (2022) Innate immune responses against the fungal pathogen Candida auris. Nat Commun 13: 3553.
Qiang Z, Sun H, Ge F, Li W, Li C, Wang S, Zhang B, Zhu L, Zhang S, Wang X, Lai J, Qin F, Zhou Y, Fu Y. (2022) The transcription factor ZmMYB69 represses lignin biosynthesis by activating ZmMYB31/42 expression in maize. Plant Physiol 189: 1916-1919.
Wang F, Cheng Z, Wang J, Zhang F, Zhang B, Luo S, Lei C, Pan T, Wang Y, Zhu Y, Wang M, Chen W, Lin Q, Zhu S, Zhou Y, Zhao Z, Wang J, Guo X, Zhang X, Jiang L, Bao Y, Ren Y, Wan J. (2022) Rice STOMATAL CYTOKINESIS DEFECTIVE2 regulates cell expansion by affecting vesicular trafficking in rice. Plant Physiol 189, 567–584.
Yi S, Zhang X, Zhang J, Ma Z, Wang R, Wu D, Wei Z, Tan Z, Zhang B, Wang M. (2022) Brittle Culm 15 mutation alters carbohydrate composition, degradation and methanogenesis of rice straw during in vitro ruminal fermentation. Front Plant Sci 13: 975456.
Zhao W, Kirui A, Deligey F, Mentink-Vigier F, Zhou Y, Zhang B, Wang T. (2021) Solid-state NMR of unlabeled plant cell walls: high-resolution structural analysis without isotopic enrichment. Biotechnol Biofuels 14: 14.
Xu W, Cheng H, Zhu S, Cheng J, Ji H, Zhang B, Cao S, Wang C, Tong G, Zhen C, Mu L, Zhou Y, Cheng Y. (2021) Functional understanding of secondary cell wall cellulose synthases in Populus trichocarpa via the Cas9/gRNA-induced gene knockouts. New Phytol 231: 1478–1495.
Zhao M, Tang S, Zhang H, He M, Liu J, Zhi H, Sui Y, Liu X, Jia G, Zhao Z, Yan J,
Zhang B, Zhou Y, Chu J, Wang X, Zhao B, Tang W, Li J, Wu C, Liu X, Dao X. (2020) DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling.
Proc Natl Acad SciUSA117: 21766–21774.
Zhao X, Ebert B, Zhang B, Liu H, Zhang Y, Zeng W, Rautengarten C, Li H, Chen X, Bacic A, Wang G, Men S, Zhou Y, Heazlewood J, Wu A. (2020) UDP-Api/UDP-Xyl synthases affect plant development by controlling the content of UDP-Api to regulate the RG-II-borate complex. Plant J 104: 252–267.
Zhao B, Tang Y, Zhang B, Wu P, Li M, Xu X, Wu G, Jiang H, Chen Y. (2019) The temperature-dependent retention of introns in GPI8 transcripts contributes to a drooping and fragile shoot phenotype in rice. Int J Mol Sci 21: 299.
Wei K, Zhao Y, Zhou H, Jiang C, Zhang B, Zhou Y, Song X, Lu M. (2019) PagMYB216 is involved in the regulation of cellulose synthesis in Populus. Mol Breeding 39: 65.
Shi Y, Liu A, Li J, Zhang J, Zhang B, Ge Q, Jamshed M, Lu Q, Li S, Xiang X, Gong J, Gong W, Shang H, Deng X, Pan J, Yuan Y. (2019) Dissecting the genetic basis of fiber quality and yield traits in interspecific backcross populations of Gossypium hirsutum x Gossypium barbadense. Mol Genet Genomics 294: 1385-1402.
He M, Lan M, Zhang B, Zhou Y, Wang Y, Zhu L, Yuan M, Fu Y. (2018) Rab-H1b is essential for trafficking of cellulose synthase and for hypocotyl growth in Arabidopsis thaliana. J Integr Plant Biol 60: 1051–1069.
Wan J, Zhu X, Wang Y, Liu L, Zhang B, Li G, Zhou Y, Zheng S. (2018) Xyloglucan fucosylation modulates arabidopsis cell wall hemicellulose aluminium binding capacity. Sci Rep 8: 428.
Zhou T, Hua Y, Zhang B, Zhang X, Zhou Y, Shi L, Xu F. (2017) Low-Boron tolerance strategies involving pectin-mediated cell wall mechanical properties in Brassica napus. Plant Cell Physiol 58: 1991–2005.
Xu Z, Li S, Zhang C, Zhang B, Zhu K, Zhou Y, Liu Q. (2017) Genetic connection between cell-wall composition and grain yield via parallel QTL analysis in indica and japonica subspecies. Sci Rep 7: 12561.
Wang X, Cheng Z, Zhao Z, Gan L, Qin R, Zhou K, Ma W, Zhang B, Wang J, Zhai H, Wan J. (2016) BRITTLE SHEATH1 encoding OsCYP96B4 is involved in secondary cell wall formation in rice. Plant Cell Rep 35: 745-755.
Shi Y, Zhang B, Liu A, Li W, Li J, Lu Q, Zhang Z, Li S, Gong W, Shang H, Gong J, Chen T, Ge Q, Wang T, Zhu H, Liu Z, Yuan Y. (2016) Quantitative trait loci analysis of Verticillium wilt resistance in interspecific backcross populations of Gossypium hirsutum x Gossypium barbadense. BMC Genomics 17: 877.
Ma J, Cheng Z, Chen J, Shen J, Zhang B, Ren Y, Ding Y, Zhou Y, Zhang H, Zhou K, Wang J, Lei C, Zhang X, Guo X, Gao H, Bao Y, Wan J. (2016) Phosphatidylserine synthase controls cell elongation especially in the uppermost internode in rice by regulation of exocytosis. PLoS ONE 11: e0153119.
Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, Zhou Y. (2015) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27: 1681-1696.
Wang X, Jing Y, Zhang B, Zhou Y, Lin R. (2015) Glycosyltransferase-like protein ABI8/ELD1/KOB1 promotes Arabidopsis hypocotyl elongation through regulating cellulose biosynthesis. Plant Cell Environ 38: 411–422.
Shi Y, Li W, Li A, Ge R, Zhang B, Li J, Liu G, Li J, Liu A, Shang H, Gong J, Gong W, Yang Z, Tang F, Liu Z, Zhu W, Jiang J, Yu X, Wang T, Wang W, Chen T, Wang K, Zhang Z, Yuan Y. (2015) Constructing a high-density linkage map for Gossypium hirsutum x Gossypium barbadense and identifying QTLs for lint percentage. J Integr Plant Biol 57: 450-467.
Zhu X, Sun Y, Zhang B, Mansoori N, Wan J, Liu Y, Wang Z, Shi Y, Zhou Y, Zheng S. (2014) TRICHOME BIREFRINGENCE-LIKE27 affects aluminum sensitivity by modulating the O-acetylation of xyloglucan and aluminum-binding capacity in Arabidopsis. Plant Physiol 166: 181–189.
Wang C, Li S, Ng S, Zhang B, Zhou Y, Whelan J, Wu P, Shou H. (2014) Mutation in xyloglucan 6-xylosytransferase results in abnormal root hair development in Oryza sativa. J Exp Bot 65: 4149-4157.
Song X, Liu L, Jiang Y, Zhang B, Gao Y, Liu X, Lin Q, Ling H, Zhou Y. (2013) Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol Plant 6: 768-780.
Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, Zhou Y. (2013) Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9: e1003704.
Kang J, Zhang H, Sun, T, Shi Y, Wang J, Zhang B, Wang Z, Zhou Y, Gu H. (2013) Natural variation of C-repeat-binding factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytol 199: 1069–1080.
Zou H, Zhang Y, Wei W, Chen H, Song Q, Liu Y, Zhao M, Wang F, Zhang B, Lin Q, Zhang W, Ma B, Zhou Y, Zhang J, Chen S. (2013) The transcription factor AtDOF4.2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem J 449: 373-388
Zhu X, Shi Y, Lei G, Fry S, Zhang B, Zhou Y, Braam J, Jiang T, Xu X, Mao C, Pan Y, Yang J, Wu P, Zheng S. (2012) XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 24: 4731–4747.
Zhang S, Song X, Yu B, Zhang B, Sun C, Knox P, Zhou Y. (2012) Identification of quantitative trait loci affecting hemicellulose characteristics based on cell wall composition in a wild and cultivated rice species. Mol Plant 5: 162-175.
Bao C, Wang J, Zhang R, Zhang B, Zhang H, Zhou Y, Huang S. (2012) Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. Plant J 71: 962-975.
Chen M, Zhang A, Zhang Q, Zhang B, Nan J, Li X, Liu N, Qu H, Lu C, Sudmorgen, Zhou Y, Xu Z, Bai S. (2012) Arabidopsis NMD3 is required for nuclear export of 60S ribosomal subunits and affects secondary cell wall thickening. PLoS ONE 7: e35904.
Lei M, Liu Y, Zhang B, Zhao Y, Wang X, Zhou Y, Raghothama KG, Liu D. (2011) Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol 156: 1116–1130.
Wang Z, Xie Z, Zhang B, Hou L, Zhou Y, Li L, Han X. (2011) Aerobic and anaerobic nonmicrobial methane emissions from plant material. Environ Sci Technol 45: 9531-9537.
Zhou Y, Li S, Qian Q, Zeng D, Zhang M, Guo L, Liu X, Zhang B, Deng L, Liu X, Luo G, Wang X, Li J. (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57: 446-462.
Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y. (2009) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60: 1055-1069.
参编专著
作物种业前沿科技战略研究. 邓小明, 杨维才, 张松梅, 葛毅强, 郑筱光, 田志喜 主编. 北京: 科学出版社, 2024.11 (编委)
植物细胞壁与木材形成. 林金星, 贺新强 主编. 北京: 高等教育出版社, 2023.7 (编委)
Wen Z, Li J, Zhang L, Zhang B, Zhou Y. (2024) Polysaccharides Analysis of Cell Wall Using Carbohydrate Gel Electrophoresis. In: Jiang, L., Shen, J., Gao, C., Wang, X. (eds) Plant Protein Secretion. Methods in Molecular Biology, vol 2841. Humana, New York, NY. (Chapter)
Zhang B, Zhou Y. (2017) Carbohydrate Composition Analysis in Xylem. In: de Lucas, M., Etchhells, J. (eds) Xylem. Methods in Molecular Biology, vol 1544. Humana Press, New York, NY. (Chapter)
更新时间:2025-12
(全部70余篇论著及更新可见网页https://www.researchgate.net/profile/Baocai-Zhang/research)