发表文章 (*Corresponding author; #Co-first author)
2023
1.Jinchao Chen#, Long Zhao#, Haoran Li#, Changfeng Yang, Dongzhi Wang, Xuelei Lin, Yujing Lin, Hao Zhang, Xiaomin Bie, Peng Zhao, Shengbao Xu, Xiansheng Zhang, Xueyong Zhang, Yingyin Yao,
Jun Xiao*. (2023) TaNF-Y-PRC2 orchestrates temporal control of starch and protein synthesis in wheat.
bioRxiv 2023.12.26.573020; doi: https://doi.org/10.1101/2023.12.26.573020
2.Xuemei Liu#, Xuelei Lin#*, Min Deng#, Bingxin Shi, Jinchao Chen, Haoran Li, Shujuan Xu, Xiaomin Bie, Xiansheng Zhang, Kang Chong,
Jun Xiao*. (2023) Distinct roles of H3K27me3 and H3K36me3 in vernalization response, maintenance and resetting in winter wheat.
bioRxiv 2023.12.19.572364; doi: https://doi.org/10.1101/2023.12.19.572364
3.Hao Zhang, Jinyuan Shi, Fa Cui, Long Zhao, Xiaoyu Zhang, Jinchao Chen, Jing Zhang, Yongpeng Li, Yanxiao Niu, Lei Wang, Caixia Gao, Xiangdong Fu, Yiping Tong, Hong-Qing Ling*, Junming Li*, Jun Xiao*. (2023) Epigenetic modifications mediate cultivar-specific root development and metabolic adaptation to nitrogen availability in wheat. Nat Commun. 2023 Dec 12;14(1):8238. doi: 10.1038/s41467-023-44003-6.
4.Zhang J, Zhang Z, Zhang R, Yang C, Zhang X, Chang S, Chen Q, Rossi V, Zhao L, Xiao J, Xin M, Du J, Guo W, Hu Z, Liu J, Peng H, Ni Z, Sun Q, Yao Y. Type I MADS-box transcription factor TaMADS-GS regulates grain size by stabilizing cytokinin signalling during endosperm cellularization in wheat. Plant Biotechnol J. 2023 Sep 26. doi: 10.1111/pbi.14180. Epub ahead of print. PMID: 37752705.
6.Xiaowei Lin*, Tingting Yuan, Hong Guo, Yi Guo, Nobutoshi Yamaguchi, Shuge Wang, Dongxia Zhang, Dongmei Qi, Jiayu Li, Qiang Chen, Xinye Liu,
Long Zhao,
Jun Xiao, Doris Wagner, Sujuan Cui*, Hongtao Zhao*. The regulation of chromatin configuration at AGAMOUS locus by LFR-SYD-containing complex is critical for reproductive organ development in Arabidopsis.
The Plant Journal,
https://doi.org/10.1111/tpj.16385
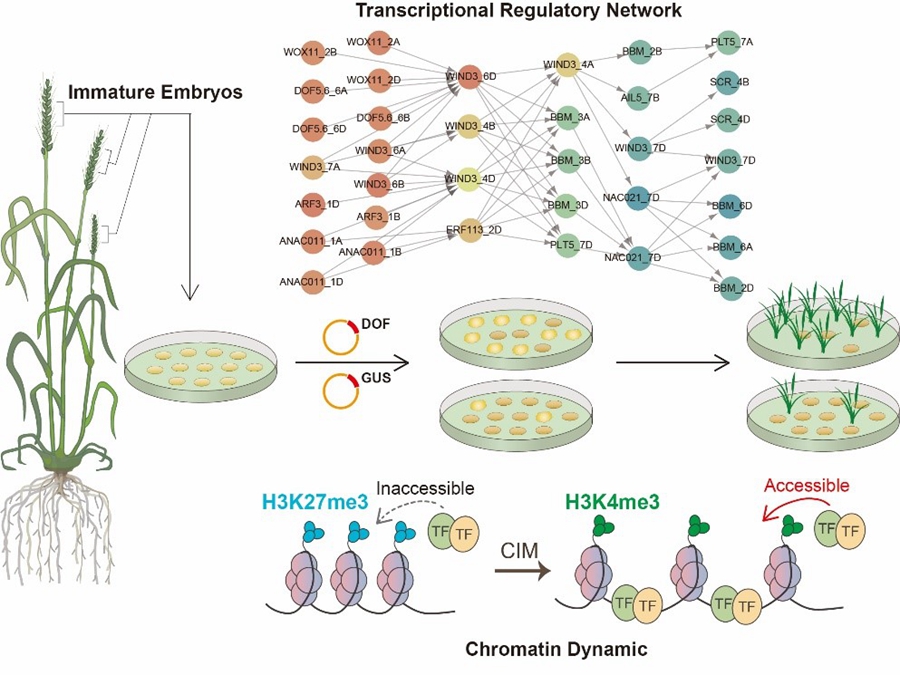
7.Xuemei Liu#, Xiaomin Bie#, Xuelei Lin#, Menglu Li, Hongzhe Wang, Xiaoyu Zhang, Yiman Yang, Chunyan Zhang, Xiansheng Zhang*, Jun Xiao*. (2022) Uncovering transcriptional regulatory network during regeneration for boosting wheat transformation. Nat Plants. 2023 May 4. doi: 10.1038/s41477-023-01406-z. Epub ahead of print. PMID: 37142750.
8.Dongzhi Wang#, Yongpeng Li#, Haojie Wang#, Yongxin Xu#, Yiman Yang#, Yuxin Zhou, Zhongxu Chen, Yuqing Zhou, Lixuan Gui, Chunjiang Zhou, Wenqiang Tang, Shuzhi Zheng, Lei Wang, Xiulin Guo, Yingjun Zhang, Fa Cui, Xuelei Lin, Yuling Jiao, Yuehui He, Junming Li*, Fei He*, Xigang Liu*, Jun Xiao* (2022) Boosting wheat functional genomics via indexed EMS mutant library of KN9204. Plant Commun. 2023 Mar 20:100593. doi: 10.1016/j.xplc.2023.100593.
9.Xuelei Lin#, Yongxin Xu#, Dongzhi Wang#, Yiman Yang, Xiaoyu Zhang, Xiaomin Bie, Hongzhe Wang, Jiafu Jiang, Zhongxu Chen, Lixuan Gui, Yiliang Ding, Yuqing Che, Aili Li, Xueyong Zhang, Fei Lu, Xiangyu Zhao, Xiansheng Zhang, Xiangdong Fu,
Jun Xiao*. (2023) Systematic mining and functional analysis of factors regulating wheat spike development for breeding selection.
BioRxiv doi: https://doi.org/10.1101/2022.11.11.516122
10. Xumei Luo#, Yiman Yang#, Xuelei Lin*, Jun Xiao*. Decipherign spike architecture formation towards yield improvement in wheat. J Genet Genomics. 2023 Mar 10:S1673-8527(23)00054-1. doi: 10.1016/j.jgg.2023.02.015.
11. Long Zhao*, Yiman Yang, Jinchao Chen, Xuelei Lin, Hao Zhang, Hao Wang, Hongzhe Wang, Xiaomin Bie, Jiafu Jiang, Xiaoqi Feng, Xiangdong Fu, , Xiansheng Zhang, Zhuo Du, Jun Xiao*. Dynamic chromatin regulatory programs during embryogenesis of hexaploid wheat. Genome Biology, 2023 Jan 13;24(1):7. doi: 10.1186/s13059-022-02844-2.
2022
1.
Jun Xiao*, Bao Liu, Yingyin Yao, Zifeng Guo, Haiyan Jia, Lingrang Kong, Aimin Zhang, Wujun Ma, Zhongfu Ni, Shengbao Xu, Fei Lu, Yuannian Jiao, Wuyun Yang,
Xuelei Lin, Silong Sun, Zefu Lu, Lifeng Gao, Guangyao Zhao, Shuanghe Cao, Qian Chen, Kunpu Zhang, Mengcheng Wang, Meng Wang, Zhaorong Hu, Weilong Guo, Guoqiang Li, Xin Ma, Junming Li, Fangpu Han, Xiangdong Fu, Zhengqiang Ma, Daowen Wang*, Xueyong Zhang*, Hong-Qing Ling*, Guangmin Xia*, YipingTong*, Zhiyong Liu*, Zhonghu He*, Jizeng Jia*, Kang Chong*. Wheat genomic study for genetic improvement of traits in China.
Science China Life Science. 2022 August, 65: 1718-1775.
https://www.sciengine.com/SCLS/doi/10.1007/s11427-022-2178-7.
2.Xiaoli Shi#, Fa Cui#, Xinyin Han#, Yilin He#,
Long Zhao#, Na Zhang#,
Hao Zhang#, Haidong Zhu, Zhexin Liu, Bin Ma, Shusong Zheng, Wei Zhang, Jiajia Liu, Xiaoli Fan, Yaoqi Si, Shuiquan Tian, Jianqing Niu, Huilan Wu,
Xuemei Liu, Zhuo Chen, Deyuan Meng, Xiaoyan Wang, Liqiang Song, Lijing Sun, Jie Han, Hui Zhao, Jun Ji, Zhiguo Wang, Xiaoyu He, Ruilin Li, Xuebin Chi, Chengzhi Liang*, Beifang Niu*,
Jun Xiao*, Junming Li*, HongQing Ling*. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen use efficiency in the wheat cultivar Kenong 9204.
Mol Plant. 2022.
https://doi.org/10.1016/j.molp.2022.07.008
3.Shengnan Li#, Dexing Lin#, Yunwei Zhang#, Min Deng#, Yongxing Chen, Bin Lv, Boshu Li, Yuan Lei, Yanpeng Wang, Long Zhao, Yueting Liang. Jinxing Liu, Kunling Chen, Zhiyong Liu, Jun Xiao*, Jinlong Qiu*, Caixia Gao*. Genome edited powdery mildew resistance in wheat without growth penalties, Nature. 2022 Feb;602(7897):455-460. doi: 10.1038/s41586-022-04395-9.
5.Tingting Yang, Dingyue Wang, Guanmei Tian, Linhua Sun, Minqi Yang, Xiaochang Yin, Jun Xiao, Yu Sheng, Danmeng Zhu, Hang He, Yue Zhou*. Chromatin remodeling complexes regulate genome architecture in Arabidopsis. Plant Cell. 2022 Jul 4;34(7):2638-2651. doi: 10.1093/plcell/koac117. PMID: 35445713; PMCID: PMC9252501.
6.Yanmin Chen, Jun Xiao, Peng Liu. Dissecting the plant chromatin interactome using mass spectrometry. Trends Biotechnol. 2022 Mar;40(3):261-265. doi: 10.1016/j.tibtech.2021.06.002.
2021
1.Tomasz Bieluszewski#, Jun Xiao#, Yiman Yang, Doris Wagner. (2021) PRC2 activity, recruitment, and silencing: a comparative perspective. Trends Plant Sci. 2021 Jul 19:S1360-1385(21)00148-5. doi: 10.1016/j.tplants.2021.06.006. Epub ahead of print. PMID: 34294542.
2.Shujuan Xu, Qi Dong, Min Deng, Dexing Lin, Jun Xiao, Peilei Cheng, Lijing Xing, Yuda Niu, Caixia Gao, Wenhao Zhang, Yunyuan Xu, Kang Chong.(2021) The vernalization-induced long non-coding RNA VAS functions with the transcription factor TaRF2b to promote TaVRN1 expression for flowering in hexaploid wheat. Mol Plant. 2021 Sep 6;14(9):1525-1538. doi: 10.1016/j.molp.2021.05.026. Epub 2021 May 27. PMID: 34052392.
3.Run Jin#, Samantha Klasfeld#, Yang Zhu, Meilin Fernandez Garcia, Jun Xiao, Soon-Ki Han, Adam Konko, Doris Wagner. LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate. Nature Communications, 2021 12(1):626. doi: 10.1038/s41467-020-20883-w.
2019
1.Bo Sun, Yingying Zhou, Jie Cai, Erlei Shang, Nobutoshi Yamaguchi,
Jun Xiao, Liang-Sheng Looi, Wan-Yi Wee, Xiuying Gao, Doris Wagner, Toshiro Ito. Integration of transcriptional repression and Polycomb-mediated silencing of WUSCHEL in floral meristems.
Plant Cell 2019. DOI:
https://doi.org/10.1105/tpc.18.00450.
2.Shujuan Xu,
Jun Xiao, Fang Yin, Xiaoyu Guo, Lijing Xing, Yunyuan Xu, Kang Chong. The protein modifications of O-GlcNAcylation and phosphorylation mediate vernalization response for flowering in winter wheat.
Plant Physiology,2019. DOI:
https://doi.org/10.1104/pp.19.00081.
2018 and before (PhD and Post-doc work)
1.Xiao J, Jin R, Yu X, Shen M, Wagner J, Pai A, Song C, Zhuang M, Kelasfeld S, He CS, Santos AM, Helliwell C, Pruneda-Paz J, Kay S, Lin XW, Cui SJ, Fernandez M, Clarenz O, Goodrich J, Zhang XY, Austin R, Bonasio R, Wagner D. (2017) Cis- and trans-determinants of Polycomb Repressive Complex 2 recruitment in Arabidopsis. Nature Genetics, doi:10.1038/ng.3937 (Nature Genetic news and views highlight, F1000 Prime)
2.Xiao J, Xu S#, Li C#, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, Gao C, Chong K. (2014) O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nature Communications 5:4572. (F1000 Prime)
3.Wu MF#, Yamaguchi N#, Xiao J#, Bargmann B, Estelle M, Sang Y, Wagner D. (2015) An auxin-regulated chromatin state switch directs organogenesis in Arabidopsis. eLife 4:e09269. (Co-first author, F1000 Prime)
4.Xiao J#, Jin R#, Wagner D. (2017) Developmental transitions: integrating environmental cues with hormonal signaling in the chromatin landscape. Genome Biology, 18 (1):88.
5.Xiao J, Wagner D. (2015) Polycomb repression in the regulation of growth and development in Arabidopsis. Curr Opin Plant Biol 23C:15-24.
6.Xiao J#, Li CH#, Xu SJ, Xing LJ, Xu YY, Chong K. (2015) AtJAC1 Regulates Nuclear Accumulation of GRP7, influencing RNA processing of FLC antisense transcripts and flowering time in Arabidopsis. Plant Physiol 169 (3):2102-2117.
7.Xiao J, Lee US, Wagner D. (2016) Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr Opin Plant Biol 4:41-53.
8.Xiao J, Zhang H, Xing LJ, Xu SJ, Liu HH, Chong K, Xu YY. (2013) Requirement of histone acetyltransferases HAM1/2 for epigenetic modification of FLC to regulate flowering in Arabidopsis. J Plant Physiol 170(4):444-451.
9.Xing LJ, Liu Y, Xu SJ, Xiao J, Wang B, Deng HW, Lu Z, Xu YY, Chong K. (2018) Arabidopsis O-GlcNAc transferase SEC actives histone methyltransferase ATX1 to regulate flowering. EMBO J, doi:10.15252/embj.201798115.
10. Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Xiao J, Guo X, Xu S, Niu Y, Jin J, Zhang H, Xu X, Li L, Wang W, Qian Q, Ge S, Chong K. (2015) COLD1 Confers Chilling Tolerance in Rice. Cell 160(6):1209-1221. (F1000 Prime, Cover story)
11. Bossi F, Fan J, Xiao J, Chandra L, Shen M, Wagner D, Rhee SY. (2017) Systematic discovery of novel eukaryotic transcriptional regulators using non-sequence homology based prediction. BMC Genomics, 18:480 DOI: 10.1186/s12864-017-3853-9
12. Liu YJ, Xu YY, Xiao J, Ma QB, Li D, Xue Z, Chong K. (2011) OsDOG, a gibberellin-induced A20/AN1 zinc-finger protein, negatively regulates gibberellin-mediated cell elongation in rice. J Plant Physiol 168(10):1098-1105.
13. Li J, Jiang JF, Qian Q, Xu YY, Zhang C, Xiao J, Du C, Luo W, Zou GX, Chen ML, Huang YQ, Feng YQ, Cheng ZK, Yuan M, Chong K. (2011) Mutation of Rice BC12/GDD1, Which Encodes a Kinesin-Like Protein That Binds to a GA Biosynthesis Gene Promoter, Leads to Dwarfism with Impaired Cell Elongation. Plant Cell 23(2):628-640.
14. Ma QB, Dai XY, Xu YY, Guo J, Liu YJ, Chen N, Xiao J, Zhang DJ, Xu ZH, Zhang XS* and Chong K*. (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150: 244-256.
软件著作权及专利
2023SR1031506
Find. Cis. Regulatory. Element. Target软件【简称:FCT软件】V1.0
HC221882 TaRF1基因及其编码的蛋白在提高小麦转化效率中的应用(3/7)
HC221883 TaDOF5.6基因在小麦高效遗传转化的应用(4/7)