RECENT MAJOR PUBICATIONS
(see http://plantbiol.genetics.ac.cn/yjcg/ for a complete list)
Research Papers:
1. Zhang, H., Zhou, L., Zhao, H., Liang, J., Liu, Y., Wang, C., Sun, S., Song, L., Zhang, Y., Cheng, Y.*, and Xue, Y.* (2025). Chromosome-scale haplotype-resolved genome assembly of the autotetraploid alfalfa cultivar Bolivia. Plant Biotechnol J 23, 4773-4775. 10.1111/pbi.70259.
2. Cai, Z., Su, Y., Kong, L., Ren, G., Zheng, L., Zhu, M., Qiu, B., Hong, Z., Ye, Y., Xue, Y., Wu W. and Duan, Y. (2025). JAGGED and DROOPING LEAF synergistically establish sexual organs and terminate floral meristem in rice. Plant J 123, e70346. 10.1111/tpj.70346.
3. Wang 王晨, C., Zhao 赵洪, H., Zhang 张洪魁, H., Sun 孙思杰, S., and Xue 薛勇彪, Y.* (2025). PSIA: A Comprehensive Knowledgebase of Plant Self-incompatibility. Genomics Proteomics Bioinformatics 23. 10.1093/gpbjnl/qzaf046.
4. Cai, Z., Li, J., Su, Y., Zheng, L., Zhang, J., Zhu, M., Qiu, B., Kong, L., Ye, Y., Xue, Y., Wu W. and Duan, Y. (2025). The MADS6, JAGGED, and YABBY proteins synergistically determine floral organ development in rice. Plant Physiol 197. 10.1093/plphys/kiaf076.
5. Chi, L., Dong, Y., Wang, R., Yang, S., Wu, L., Xue, Y.* and Chen, H.* (2025) HapNetworkView: a tool for haplotype network exploration and visualization. BMC Genomics. 26(1):52.
6. Li, L., Li, C., Li, N., Zou, D., Zhao, W., Luo, H., Xue, Y., Zhang, Z., Bao, Y., Song, S. (2024) Machine Learning Early Detection of SARS-CoV-2 High-Risk Variants. Adv. Sci. (Weinh). 11(45): e2405058
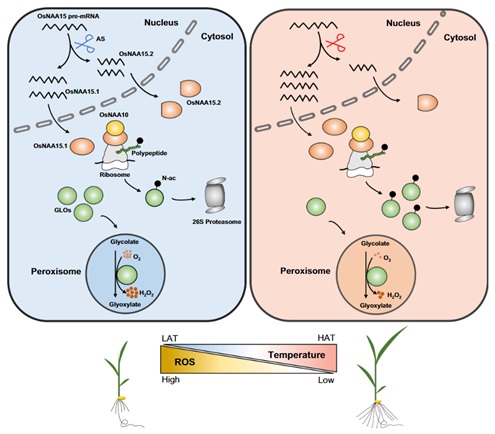
7. Li, X., Tang, H., Xu, T., Wang, P., Ma, F., Wei, H., Fang, Z., Wu, X., Wang, Y., Xue, Y., Zhang, B. (2024) N-terminal acetylation orchestrates glycolate-mediated ROS homeostasis to promote rice thermoresponsive growth. New Phytol. 243(5):1742-1757
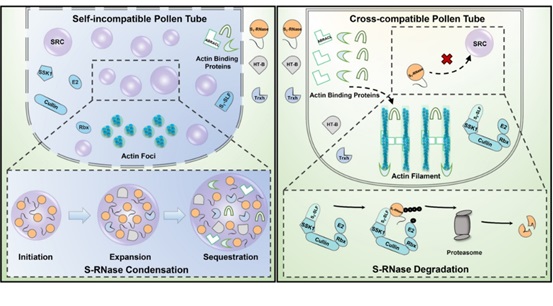
8. Tian, H., Zhang, H., Huang, H., Zhang, Y., and Xue, Y. (2024). Phase separation of S-RNase promotes self-incompatibility in Petunia hybrida. J. Integr. Plant. Biol. 66, 986-1006.
9. Zhu, S., Zhang, Y., Copsy, L., Han, Q., Zheng, D., Coen, E., and Xue, Y. (2023). The snapdragon genomes reveal the evolutionary dynamics of the S-locus supergene. Mol. Biol. Evol. 40, msad080.
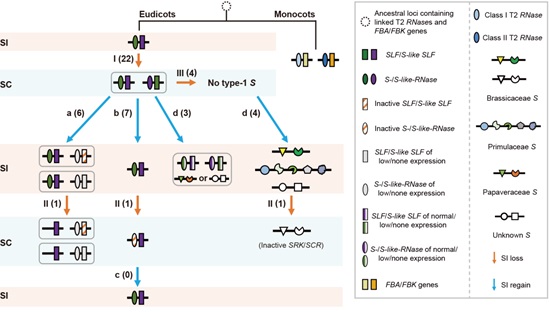
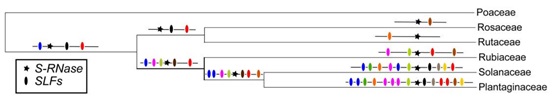
10. Zhao, H., Zhang, Y., Zhang, H., Song, Y., Zhao, F., Zhang, Y., Zhu, S., Zhang, H., Zhou, Z., Guo, H., Li, M., Li, J., Gao, Q., Han, Q., Huang, H., Copsey, L., Li, Q., Chen, H., Coen, E., Zhang, Y. and Xue, Y. (2022) Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596
11. CNCB-NGDC Members and Partners (2022). Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Research, 50(D1), D27–D38.
12. Zhang, Y., Li, Z., Zhang, Y., Lin, K., Peng, Y., Ye, L., Zhuang, Y., Wang, M., Xie, Y., Guo, J., Teng, W., Tong, Y., Zhang, W., Xue, Y., Lang, Z. and Zhang, Y. (2021). Evolutionary rewiring of the wheat transcriptional regulatory network by lineage-specific transposable elements. Genome Research, 31(12), 2276–2289.
13. Liu, Q., Du, Z., Zhu, S., Zhao, W., Chen, H. and Xue, Y. (2021). Metagenomic evidence for the co-existence of SARS and H1N1 in patients from 2007-2012 flu seasons in France. Biosafety and Health.
14. CNCB-NGDC Members and Partners (2021). Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Research, 49(D1), D18–D28.
15. Wu, C. I., Wen, H., Lu, J., Su, X. D., Hughes, A. C., Zhai, W., Chen, C., Chen, H., Li, M., Song, S., Qian, Z., Wang, Q., Chen, B., Guo, Z., Ruan, Y., Lu, X., Wei, F., Jin, L., Kang, L., Xue, Y., … Zhang, Y. P. (2021). On the origin of SARS-CoV-2-The blind watchmaker argument. Science China. Life sciences, 64(9), 1560–1563.
16. Zhao, S., Sha, T., Xue, Y., Wu, C. I., & Chen, H. (2021). Will the large-scale vaccination succeed in containing the covid-19 epidemic and how soon? Quantitative Biology.
17. Wang, Q., Chen, H., Shi, Y., Hughes, A. C., Liu, W. J., Jiang, J., Gao, G. F., Xue, Y., & Tong, Y. (2021). Tracing the origins of SARS-CoV-2: lessons learned from the past. Cell Research, 31(11), 1139–1141.
18. Wang, M., Li, Z., Zhang, Y., Zhang, Y., Xie, Y., Ye, L., Zhuang, Y., Lin, K., Zhao, F., Guo, J., Teng, W., Zhang, W., Tong, Y., Xue, Y., & Zhang, Y. (2021). An atlas of wheat epigenetic regulatory elements reveals subgenome divergence in the regulation of development and stress responses. The Plant Cell, 33(4), 865–881.
19. Zhao, H., Song, Y., Li, J., Zhang, Y., Huang, H., Li, Q., Zhang, Y., & Xue, Y. (2021). Primary restriction of S-RNase cytotoxicity by a stepwise ubiquitination and degradation pathway in Petunia hybrida. New Phytologist, 231(3), 1249–1264.
20. Song, S., Ma, L., Zou, D., Tian, D., Li, C., Zhu, J., Chen, M., Wang, A., Ma, Y., Li, M., Teng, X., Cui, Y., Duan, G., Zhang, M., Jin, T., Shi, C., Du, Z., Zhang, Y., Liu, C., Li, R., … Xue, Y., Bao, Y. (2020). The Global Landscape of SARS-CoV-2 Genomes, Variants, and Haplotypes in 2019nCoVR. Genomics, Proteomics & Bioinformatics, 18(6), 749–759.
21. Gong, Z., Zhu, J. W., Li, C. P., Jiang, S., Ma, L. N., Tang, B. X., Zou, D., Chen, M. L., Sun, Y. B., Song, S. H., Zhang, Z., Xiao, J. F., Xue, Y., Bao, Y. M., Du, Z. L., & Zhao, W. M. (2020). An online coronavirus analysis platform from the National Genomics Data Center. Zoological Research, 41(6), 705–708.
22. Li, X., Chen, Z., Zhang, G., Lu, H., Qin, P., Qi, M., Yu, Y., Jiao, B., Zhao, X., Gao, Q., Wang, H., Wu, Y., Ma, J., Zhang, L., Wang, Y., Deng, L., Yao, S., Cheng, Z., Yu, D., Zhu, L., Xue, Y., … Liang, C. (2020). Analysis of genetic architecture and favorable allele usage of agronomic traits in a large collection of Chinese rice accessions. Science China. Life Sciences, 63(11), 1688–1702.
23. CNCB-NGDC Members and Partners (2021). Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2021. Nucleic Acids Research, 49(D1), D18–D28.
24. Zhao, W., Qu, X., Zhuang, Y., Wang, L., Bosch, M., Franklin-Tong, V. E., Xue, Y., & Huang, S. (2020). Villin controls the formation and enlargement of punctate actin foci in pollen tubes. Journal of Cell Science, 133(6), jcs237404.
25. Liu, Q., Zhao, S., Shi, C. M., Song, S., Zhu, S., Su, Y., Zhao, W., Li, M., Bao, Y., Xue, Y., & Chen, H. (2020). Population Genetics of SARS-CoV-2: Disentangling Effects of Sampling Bias and Infection Clusters. Genomics, Proteomics & Bioinformatics, 18(6), 640–647.
26. Zhao, W. M., Song, S. H., Chen, M. L., Zou, D., Ma, L. N., Ma, Y. K., Li, R. J., Hao, L. L., Li, C. P., Tian, D. M., Tang, B. X., Wang, Y. Q., Zhu, J. W., Chen, H. X., Zhang, Z., Xue, Y., & Bao, Y. M. (2020). The 2019 novel coronavirus resource. Yi chuan = Hereditas, 42(2), 212–221.
27. Yarra, R., & Xue, Y. (2020). Ectopic expression of nucleolar DEAD-Box RNA helicase OsTOGR1 confers improved heat stress tolerance in transgenic Chinese cabbage. Plant Cell Reports, 39(12), 1803–1814.
28. Li, Z., Wang, M., Lin, K., Xie, Y., Guo, J., Ye, L., Zhuang, Y., Teng, W., Ran, X., Tong, Y., Xue, Y., Zhang, W., & Zhang, Y. (2019). The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biology, 20(1), 139.
29. Li, M., Zhang, D., Gao, Q., Luo, Y., Zhang, H., Ma, B., Chen, C., Whibley, A., Zhang, Y., Cao, Y., Li, Q., Guo, H., Li, J., Song, Y., Zhang, Y., Copsey, L., Li, Y., Li, X., Qi, M., Wang, J., … Xue, Y. (2019). Genome structure and evolution of Antirrhinum majus L. Nature Plants, 5(2), 174–183.
30. Pervaiz, N., Shakeel, N., Qasim, A., Zehra, R., Anwar, S., Rana, N., Xue, Y., Zhang, Z., Bao, Y., & Abbasi, A. A. (2019). Evolutionary history of the human multigene families reveals widespread gene duplications throughout the history of animals. BMC Evolutionary Biology, 19(1), 128.
31. 薛勇彪.(2018).面向国家重大需求打造国际一流研究所——庆祝中国科学院北京基因组研究所成立15周年. 遗传(11),925-928.
32. BIG Data Center Members (2019). Database Resources of the BIG Data Center in 2019. Nucleic Acids Research, 47(D1), D8–D14.
33. Zhang, Z., Zhang, B., Chen, Z., Zhang, D., Zhang, H., Wang, H., Zhang, Y., Cai, D., Liu, J., Xiao, S., Huo, Y., Liu, J., Zhang, L., Wang, M., Liu, X., Xue, Y., Zhao, L., Zhou, Y., & Chen, H. (2018). A PECTIN METHYLESTERASE gene at the maize Ga1 locus confers male function in unilateral cross-incompatibility. Nature Communications, 9(1), 3678.
34. Xue, Y. (2018). The Chinese garden of genetics -celebrating 40th anniversary of Genetics Society of China. Journal of Genetics and Genomics = Yi chuan xue bao, 45(1), 1.
35. Tavares, H., Whibley, A., Field, D. L., Bradley, D., Couchman, M., Copsey, L., Elleouet, J., Burrus, M., Andalo, C., Li, M., Li, Q., Xue, Y., Rebocho, A. B., Barton, N. H., & Coen, E. (2018). Selection and gene flow shape genomic islands that control floral guides. Proceedings of the National Academy of Sciences of the United States of America, 115(43), 11006–11011.
36. Qu, X., Zhang, R., Zhang, M., Diao, M., Xue, Y., & Huang, S. (2017). Organizational Innovation of Apical Actin Filaments Drives Rapid Pollen Tube Growth and Turning. Molecular Plant, 10(7), 930–947.
37. Bradley, D., Xu, P., Mohorianu, I. I., Whibley, A., Field, D., Tavares, H., Couchman, M., Copsey, L., Carpenter, R., Li, M., Li, Q., Xue, Y., Dalmay, T., & Coen, E. (2017). Evolution of flower color pattern through selection on regulatory small RNAs. Science (New York, N.Y.), 358(6365), 925–928.
38. Li, J., Zhang, Y., Song, Y., Zhang, H., Fan, J., Li, Q., Zhang, D., and Xue, Y. (2017) Electrostatic potentials of the S-locus F-box proteins contribute to the pollen S specificity in self-incompatibility in Petunia hybrida. Plant J 89, 45-57.
39. Zhang, B., Wu,S., Zhang Y., Xu, T., Guo, F., Tang, H., Li, X., Wang, P., Qian, W., Xue, Y.(2016)A High Temperature-Dependent Mitochondrial Lipase EXTRA GLUME1 Promotes Floral Phenotypic Robustness against Temperature Fluctuation in Rice (Oryza sativa L.). PLOS Genetics.
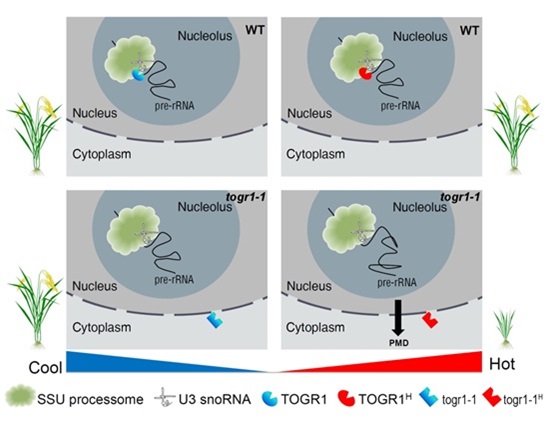
40. Wang, D., Qin, B., Li, X., Tang, D., Zhang, Y., Zheng, Z., Cheng, Z., Xue, Y.(2016) Nucleolar DEAD-Box RNA Helicase TOGR1 Regulates Thermotolerant Growth as a Pre-rRNA Chaperone in Rice. PLOS Genetics.
41. Li Q., Zhao T., Yue M., Xue, Y., Bao S. (2016)The Protein Arginine Methylase 5 (PRMT5/SKB1) Gene Is Required for the Maintenance of Root Stem Cells in Response to DNA Damage. JGG 43 (2016) 187e197
42. Liu, W., Fan, J., Li, J., Song, Y., Li,Q., Zhang, Y., Xue, Y. (2014) SCFSLF-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petuniahybrida. Front.Genet. 5:288 doi: 10.3389/fgene.2014.00228
43. Hu, X., Qian Q., Xu T., Zhang Y., Dong G., Gao, T., Xie, Q., Xue, Y. (2013) The U-Box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G a subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet 9(3): e1003391
44. Xu, C., Li, M., Wu, J., Guo, H., Li, Q., Zhang, Y., Chai, J., Li, T., Xue, Y. (2013) Identification of a canonical SCFSLF complex involved in S-RNase-based self-incompatibility of Pyrus (Rosaceae). Plant Mol. Biol. 81:245-257
45. Duan,Y. , Xing, Z., Diao, Z., Xu, W., Li, S., Du, X., Wu, G., Wang, C., Lan, T., Meng, Z., Liu, H., Wang, F., Wu, W., Xue, Y. (2012) Characterization of Osmads6-5, a null allele, reveals that OsMADS6 is a critical regulator for early flower development in rice (Oryza sativa L.) . Plant Mol. Biol. 80:429-442
46. Duan, Y., Li, S., Chen, Z., Zheng, L., Diao, Z., Zhou, Y., Lan, T., Guan, H., Pan, R., Xue, Y., Wu, W. (2012). Dwarf and Deformed Flower 1, encoding an F-box protein, is essential for regulating vegetative and floral development in rice (Oryza sativa L.). Plant J. 72:829-842
47. The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635-641
48. Chen, G., Zhang, B., Liu, L., Li, Q., Zhang, Y., Xie, Q., Z., Xue, Y. (2012) Identification of a ubiquitin-binding structure in the S-locus F-box protein controlling S-RNase-based self-incompatibility. J. Genet. Genom. 39:93-102
49. Xu, W., Yang, R., Li, M., Xing, Z., Yang, W., Chen, G., Guo, H., Gong, Z., Du, Z., Zhang, Z., Hu, X., Wang, D., Qian, Q., Wang, T., Su, Z., Xue, Y. (2011) Transcriptome phase distribution analysis reveals diurnal regulated biological processes and key pathways in rice flag leaves and seedling leaves. PLoS One 6: e17613
50. Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., Yue, M., Li, Q., Zhang, Y., Xu, Y., Xue, Y., Chong, K., Bao, S. (2011) Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23:396-411
51. Wei, L., Xu, W., Deng, Z., Su, Z., Xue, Y., Wang, T. (2010) Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11:338
52. Causier, B., Castillo, R., Xue, Y., Schwarz-Sommer, Z., Davies, B. (2010) Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Current Biol. 27:2651-2664
53. Zhao, L., Huang, J., Zhao, Z., Li, Q., Sims, T.L., Xue, Y. (2010) The SKP1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J. 62:52-63
54. Wang, K., Tang, D., Hong, L., Xu, W., Huang, J., Li, M., Gu, M., Xue, Y., Cheng, Z. (2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6(1): e1000818
55. Liu, F., Xu, W., Wei, Q., Zhang, Z., Tan, L., Chao, D., Yao, D., Wang, C., Yan, H., Sun C. Xue, Y., Su, Z. (2010) Gene expression profiles deciphering rice phenotypic variation between Nipponbare (Japonica) and 93-11(Indica) during oxidative stress. PLoS One 5(1): e8632
56. Li, H., Xue, D., Gao, Z., Yan, M., Xu, W., Xing, Z., Huang, D., Qian, Q., Xue, Y. (2009) A putative lipase gene EXTRA GLUME1 regulates both empty glume fate and spikelet development in rice. Plant J. 57:593-60
57. Zhang, Y., Xu, W., Li, Z., Deng, X., Wu, W., Xue, Y. (2008) F-box protein DOR functions as a novel inhibitory factor for ABA-induced stomatal closure under drought stress in Arabidopsis thaliana. Plant Physiol. 148:2121-2133
58. Xu, S., Deng, Z., Chong, K., Xue, Y., Wang, T. (2008) Dynamic proteomic analysis reveals a switch between central carbon metabolism and alcoholic fermentation in Oryza sativa filling grains. Plant Physiol. 148:908-925
59. Pu, L., Li, Q., Fan, X., Yang, W., Xue, Y. (2008) A R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180:811-820
60. Chen, J., Ding, J., Ouyang, Y., Du, H., Yang, J., Cheng, K., Zhao, J., Qiu, S., Zhang, X., Yao, J., Liu, K., Wang, L., Xu, C., Li, X., Xue, Y., Xia, M., Ji, Q., Lu, J., Xu. M., Zhang, Q. (2008) A tri-allelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. USA 105:11436-11441
61. Yang, W., Kong, Z., Omo-Ikerodah, E., Xu, W., Li, Q., Xue, Y. (2008) A calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). JGG 35:531-543
62. Yang, W., Lai, Y., Li, M., Xu, W., Xue, Y. (2008) A novel C2-domain phospholipid binding protein, OsPBP1, is required for pollen fertility in rice. Mol Plant 1:770-785
63. Zhang, D., Yang, Q., Ding, Y., Cao, X., Xue, Y., Cheng, Z. (2008) Cytological characterization of the tandem repetitive sequences and their methylation status in Antirrhinum majus genome. Genomics 92:107-114
64. Li, M., Xu, W., Yang, W., Kong, Z., Xue, Y. (2007) Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigam in rice (Oryza sativa L.). Plant Physiol. 144:1797-1812
65. Yang, Q., Zhang, D., Li, Q., Cheng, Z., Xue, Y. (2007) Heterochromatic and genetic features are consistent with recombination suppression of the self-incompatibility (S)-locus in Antirrhinum. Plant J. 51:140-151
66. Wang, X, Zhang, Y., Ma, Q., Zhang, Z., Xu, Z., Xue, Y., Bao, S., Chong, K. (2007) SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26:1934-1941
67. Dai, S., Chen, T., Chong, K., Xue, Y., Li, S., Wang, T. (2007) Proteomic identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol. Cell. Proteomics 6:207-230
68. Kong, Z., Li, M., Yang, W., Xu, W., Xue, Y. (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice (Oryza sativa L.). Plant Physiol. 141:1376-1388
69. Huang, J., Zhao, L., Yang, Q., Xue, Y. (2006) AhSSK1, a novel SKP1-like protein that interacts with the S-Locus F-box protein. Plant J. 46:780-793
70. Dai, S., Li, L., Chen, T., Chong, K., Xue, Y., Wang, T. (2006) Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated potentially with pollen germination and tube growth. Proteomics 6:2504-2529
71. Dong, L., Wang, L., Zhang, Y., Zhang, Y., Deng, X., Xue, Y. (2006) An auxin-inducible F-box protein CEGENDUO negatively regulates auxin-mediated lateral root formation in Arabidopsis. Plant Mol Biol. 60:599-616
72. Lan, L., Li, M., Lai, Y., Xu, W., Kong, Z., Ying, K., Han, B., Xue, Y. (2005) Microarray analysis reveals similarities and variations in genetic programs involved in pollination/fertilization and stress responses in rice (Oryza sativa L.). Plant Mol Biol 59:151-164
73. Wang, Z., Liang, Y., Li, C., Xu, Y., Lan, L., Chen, C., Xu, Z., Xue, Y., Chong, K. (2005) Identification of anther development-related genes by gene expression profiling with a 10-K cDNA microarray in rice (Oryza sativa L.). Plant Mol Biol 58:721-737
74. Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., Davies, B. (2005) Evolution in action:following function in duplicated floral homeotic genes. Curr. Biol. 15:1508-1512
75. Wang, L., Dong, L., Zhang, Y. E., Zhang, Y., Wu, W., Deng, X.W., Xue, Y. (2005) Genome-wide analysis of S-locus F-box –like genes in Arabidopsis thaliana. Plant Mol Biol 56:929-945
76. Qiao, H., Wang, F., Zhao, L., Zhou, J., Lai, Z., Zhang, Y., Robbins, T., Y., Xue, Y. (2004) The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16:2307-2322
77. Lan, L., Chen, W., Lai, Y., Suo, J., Kong, Z., Li, C., Lu, Y., Zhang, Y., Zhao, X., Zhang, X., Han, B., Cheng, J., Xue, Y. (2004) Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) using a 10K cDNA microarray. Plant Mol Biol 54:471-487
78. Qiao, H., Wang, H., Zhao, L., Zhou, J., Huang, J., Zhang, Y., Xue, Y. (2004) The F-box protein AhSLF-S2 physically interacts with S-RNases that are inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16:582-295
Reviews and Book Chapters:
1. Xue, Y. (2025). S-RNase-based self-incompatibility in angiosperms: Degradation, condensation, and evolution. Plant Physiol 199. 10.1093/plphys/kiaf360.
2. Zhao, H., Xue, Y. (2024) Molecular and evolutionary mechanisms of self-incompatibility in angiosperms. Hereditas (Beijing) 46(1): 3-17.
3. Zhao, H., Song, L., Zhang, Y., Cheng Y., Xue, Y. (2022) Self-incompatibility and inbreeding depression of forage crops. Chinese Bulletin of Botany 57(6): 742-755.
4. Chen, G., Zhang, B., Zhao, Z., Sui, Z., Zhang, H., Xue, Y. (2010) “A life or death decision” for pollen tubes in S-RNase-based self-incompatibility. J. Exp. Bot. 61:2027-2037
5. Zhang, Y., Zhao, Z., Xue, Y. (2009) Roles of proteolysis in plant self-incompatibility. Ann. Rev. Plant Biol. 60:21-42
6. Zhang, Q., Li, J., Xue, Y., Han, B., Deng, X. (2008) Rice 2020: A call for an international coordinated effort in rice functional genomics. Mol Plant 1:715-719
7. Li, C., Zhao, J., Jiang, H., Geng, Y., Dai, Y., Fan, H., Zhang, D., Chen, J., Lu, F., Shi, J., Sun, S., Chen, J., Yang, X., Lu, C., Chen, C., Cheng, Z., Ling, H., Wang, Y., Xue, Y., Li, C. (2008) A snapshot of the Chinese SOL Project. JGG 35: 387-390
8. Zhang, Y., Xue, Y. (2008) Molecular biology of S-RNase-based self-incompatibility. Pages 193-215 in Franklin-Tong, V. E. (ed.) Self-Incompatibility in Flowering Plants: Evolution, Diversity and Mechanisms, Springer-Verlag
9. Wang, Y., Xue, Y. and Li, J. (2005) Towards molecular breeding and improvement of rice in China. Trends Plant Sci. 10:610-614